 Author
Author  Correspondence author
Correspondence author
Animal Molecular Breeding, 2024, Vol. 14, No. 2
Received: 06 Feb., 2024 Accepted: 18 Mar., 2024 Published: 31 Mar., 2024
This study investigates the genetic basis of coat color variations in domestic rabbits (Oryctolagus cuniculus) by analyzing several key genes associated with pigmentation, and sequences the melanocortin 1 receptor (MC1R) gene and identified four alleles, including two in-frame deletions linked to specific coat colors such as red/fawn/yellow and black. Additionally, single-nucleotide polymorphism (SNP) markers were used to explore fur color traits across different rabbit breeds, identifying genes like ASIP, MITF, and KIT that are associated with pigmentation. A frameshift mutation in the melanophilin (MLPH) gene was found to cause the dilute coat color phenotype, providing a model for human Griscelli syndrome type. Furthermore, a composite in-frame deletion in the MC1R gene was associated with the Japanese brindling coat color, suggesting complex regulatory mechanisms. A premature stop codon in the TYRP1 gene was linked to the brown coat color, highlighting its role in eumelanin production. These findings enhance our understanding of the genetic mechanisms underlying coat color variations in domestic rabbits and provide valuable insights for breeding programs.
1 Introduction
Coat color in domestic rabbits (Oryctolagus cuniculus) is a highly variable trait influenced by both genetic and environmental factors. This phenotypic diversity is not only of aesthetic and economic importance but also provides a valuable model for studying genetic mechanisms of pigmentation. Historical studies have shown that coat color can evolve rapidly in response to environmental pressures, as evidenced by significant changes in rabbit populations over relatively short periods (Stodart, 1965). The genetic basis of these variations is complex, involving multiple genes and their interactions. Key pigmentation genes such as MC1R, MITF, TYR, TYRP1, and MLPH have been identified as major contributors to coat color diversity in rabbits (Jia et al.,, 2021).
Understanding the genetic underpinnings of coat color in rabbits is crucial for several reasons. It has direct implications for the rabbit breeding industry, where specific coat colors are often preferred for commercial purposes, such as in the Rex rabbit industry (Zhang et al., 2023). Studying these genetic variations can provide insights into the broader mechanisms of pigmentation, which are relevant to other species, including humans. Additionally, identifying genetic markers associated with coat color can aid in the conservation and management of rabbit breeds, ensuring the preservation of genetic diversity (Jia et al., 2021; Zhang et al., 2023).
This study conducts a comprehensive genetic analysis of coat color variation in domestic rabbits, identifying and characterizing genetic variations associated with different coat colors across various breeds; using genome-wide association studies (GWAS) and sequencing technologies, it identifies specific single nucleotide polymorphisms (SNPs) and genomic regions that contribute to coat color diversity, ultimately aiming to deepen our understanding of the genetic architecture of rabbit coat color and provide valuable molecular markers for breeding programs.
2 Genetic Basis of Coat Color
2.1 Overview of pigmentation genes in mammals
Pigmentation in mammals is primarily controlled by a set of key genes that regulate the production, distribution, and type of melanin produced in the skin and hair follicles. The two main types of melanin are eumelanin, which is black or brown, and pheomelanin, which is red or yellow. The balance and distribution of these pigments result in the wide variety of coat colors observed in mammals.
Several genes are crucial in the pigmentation process, including the melanocortin 1 receptor (MC1R), microphthalmia-associated transcription factor (MITF), tyrosinase (TYR), tyrosinase-related protein 1 (TYRP1), and melanophilin (MLPH). These genes interact in complex pathways to regulate melanin synthesis and distribution. For instance, MC1R is a critical receptor that influences the type of melanin produced by melanocytes. When activated, MC1R promotes the production of eumelanin over pheomelanin, leading to darker pigmentation (Fontanesi et al., 2006; Jia et al., 2021).
MITF is a transcription factor that regulates the expression of several melanogenic enzymes, including TYR and TYRP1. TYR is the enzyme responsible for the first step in melanin synthesis, converting tyrosine to dopaquinone, which then undergoes further reactions to form either eumelanin or pheomelanin. TYRP1 assists in the stabilization and function of TYR, and mutations in these genes can lead to various pigmentation disorders (Utzeri et al., 2021; Jia et al., 2021).
MLPH is involved in the transport of melanosomes, the organelles that store melanin, to the tips of melanocyte dendrites, where they are transferred to keratinocytes. Mutations in MLPH can result in diluted coat colors due to the improper distribution of melanosomes (Fontanesi et al., 2014; Demars et al., 2018).
2.2 Key genes involved in rabbit coat color variations
In domestic rabbits (Oryctolagus cuniculus), several genes have been identified as key players in determining coat color variations. These include MC1R, MITF, TYR, TYRP1, MLPH, and the agouti signaling protein (ASIP) gene.
The MC1R gene has been extensively studied in rabbits, with several alleles identified that correlate with different coat colors. For example, a 30-nucleotide in-frame deletion in MC1R is associated with red/fawn/yellow coat colors, while a 6-nucleotide in-frame deletion is linked to black coat colors (Fontanesi et al., 2006). These mutations affect the receptor's ability to bind to its ligand, leading to variations in melanin production.
The TYR gene, responsible for the initial step in melanin synthesis, has also been linked to coat color variations in rabbits. Mutations in TYR can lead to albinism or other pigmentation phenotypes. For instance, specific missense mutations in TYR are associated with the albino, Himalayan, and chinchilla phenotypes in rabbits (Utzeri et al., 2021).
MLPH mutations are known to cause the dilute coat color phenotype in rabbits. A frameshift mutation in the MLPH gene has been identified as the cause of the dilute (d) allele, which results in a lighter coat color due to the improper distribution of melanosomes (Fontanesi et al., 2014; Demars et al., 2018).
The ASIP gene plays a role in the distribution of eumelanin and pheomelanin, with different alleles leading to various coat color patterns. For example, the wild-type allele (A) results in a light-bellied agouti phenotype, while other alleles can lead to black non-agouti or black and tan phenotypes (Yang et al., 2015; Zhang et al., 2023).
2.3 Molecular mechanisms of color expression
The molecular mechanisms underlying coat color expression in rabbits involve complex interactions between various genes and their regulatory elements. These mechanisms include gene mutations, epigenetic modifications, and gene expression regulation.
Mutations in key pigmentation genes can lead to changes in the structure and function of the proteins they encode, resulting in altered melanin production and distribution. For example, the frameshift mutation in the MLPH gene causes a loss of function, leading to the dilute coat color phenotype due to the improper transport of melanosomes (Fontanesi et al., 2014; Demars et al., 2018). Similarly, mutations in the MC1R gene can affect the receptor's ability to promote eumelanin production, leading to variations in coat color (Fontanesi et al., 2006).
Epigenetic modifications, such as DNA methylation, also play a significant role in coat color expression. A study on Rex rabbits revealed that DNA methylation patterns in hair follicles are associated with inherited color dilution. Differentially methylated regions (DMRs) were identified between rabbits with different coat colors, suggesting that epigenetic regulation can influence pigmentation by altering gene expression (Chen et al., 2020).
Gene expression regulation is another critical aspect of coat color determination. Transcriptome analysis of rabbit skin tissues has shown that the expression levels of key pigmentation genes, such as TYR and TYRP1, vary between different coat color phenotypes. This differential expression can result in the production of different amounts and types of melanin, leading to the observed coat color variations (Qin et al., 2016).
3 Common Coat Color Variants in Domestic Rabbits
3.1 Description of major coat colors
Domestic rabbits exhibit a variety of coat colors, with some of the most common being albino, agouti, and black. The albino variant is characterized by a complete lack of pigmentation, resulting in white fur and red eyes due to the visibility of blood vessels in the absence of melanin. This phenotype is often associated with mutations in the tyrosinase (TYR) gene, which is crucial for melanin production (Utzeri et al., 2021). The agouti coat color, on the other hand, features a banded pattern of pigmentation, where each hair has multiple colors, typically with a dark base, a lighter middle band, and a dark tip. This pattern is regulated by the agouti signaling protein (ASIP) gene, which influences the distribution of eumelanin and pheomelanin along the hair shaft (Zhang et al., 2023). The black coat color is a result of high eumelanin production, giving the fur a uniform dark appearance. This phenotype is often linked to the melanocortin 1 receptor (MC1R) gene, which plays a significant role in the regulation of melanin synthesis (Fontanesi et al., 2006).
3.2 Genetic mutations responsible for each variant
The genetic basis for these coat color variants involves specific mutations in key pigmentation genes. For the albino phenotype, mutations in the TYR gene are critical. Disruptive mutations in TYR, such as missense mutations p.T373K and p.E294G, impair the enzyme's function, leading to a lack of melanin production and resulting in albinism (Utzeri et al., 2021). The agouti coat color is primarily influenced by the ASIP gene. Variations in this gene, particularly those affecting its expression or function, result in the characteristic banded hair pattern seen in agouti rabbits (Zhang et al., 2023). The black coat color is associated with mutations in the MC1R gene. Specific alleles, such as the c.280_285del6 deletion, have been identified in black-coated rabbits, which enhance the receptor's activity, leading to increased eumelanin production (Fontanesi et al., 2006). Additionally, the melanophilin (MLPH) gene has been implicated in coat color dilution, where mutations such as the c.585delG frameshift mutation result in a diluted pigmentation phenotype, affecting both eumelanin and pheomelanin.
3.3 Inheritance patterns of coat colors
The inheritance patterns of these coat color variants follow Mendelian principles, with some traits being dominant and others recessive. The albino phenotype, caused by mutations in the TYR gene, is typically inherited in an autosomal recessive manner. This means that two copies of the mutant allele are required for the albino phenotype to be expressed (Table 1) (Utzeri et al., 2021). The agouti coat color, influenced by the ASIP gene, follows a more complex inheritance pattern due to its interaction with other pigmentation genes. However, it is generally considered a dominant trait, where a single copy of the agouti allele can produce the banded hair pattern (Zhang et al., 2023). The black coat color, associated with the MC1R gene, can be inherited as a dominant trait when the c.280_285del6 allele is present. This allele is dominant over other MC1R alleles, such as the c.304_333del30 allele, which is associated with red/fawn/yellow coat colors (Fontanesi et al., 2006). The dilution of coat color, linked to the MLPH gene, is inherited in an autosomal recessive manner, requiring two copies of the mutant allele for the diluted phenotype to manifest (Fontanesi et al., 2014; Demars et al., 2018).
 Table 1 Polymorphisms identified in the rabbit TYR gene (Adopted from Utzeri et al., 2021) |
4 Recent Advances in Genetic Research
4.1. Genome-wide association studies (GWAS) on coat color
Genome-wide association studies (GWAS) have significantly advanced our understanding of the genetic basis of coat color variations in domestic rabbits. These studies involve scanning the entire genome to identify genetic variants associated with specific traits, such as coat color. One notable study focused on Chinese Rex rabbits, where researchers collected blood samples from 250 rabbits with six different coat colors and performed genome sequencing using a restriction site-associated DNA sequencing approach. They identified 91 546 single nucleotide polymorphisms (SNPs) distributed among 21 autosomes. The GWAS revealed 24 significant SNPs located within a genomic region on chromosome 4 (OCU4), with the agouti signaling protein (ASIP) gene identified as a putative causal gene affecting coat color variation (Zhang et al., 2023).
Another study investigated the genetic polymorphisms in pigmentation genes such as MC1R, MITF, TYR, TYRP1, and MLPH in four Chinese native rabbit breeds with different coat colors. The researchers discovered 14 genetic variants with low-to-moderate polymorphism and significant differences in gene frequency among the rabbit populations. These findings suggest that these genetic variations play a crucial role in regulating coat color in rabbits (Jia et al., 2021).
4.2 CRISPR and other genetic editing techniques
The advent of CRISPR/Cas9 and other genetic editing techniques has opened new avenues for manipulating coat color in domestic rabbits. A groundbreaking study utilized the CRISPR/Cas9 system to induce mutations in the melanocortin 1 receptor (MC1R) gene, resulting in a novel pale-yellow coat color in rabbits. The researchers designed two single-guide RNAs (sgRNAs) for the MC1R gene and confirmed the editing efficiency by injecting rabbit zygotes. The edited rabbits exhibited a loss of eumelanin synthesis, leading to the pale-yellow coat color. This study demonstrated the potential of gene editing technology to create novel phenotypes in rabbit breeding (Figure 1) (Xiao et al., 2019).
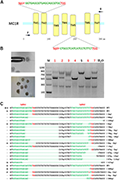 Figure 1 Dual single-guide RNA (sgRNA)-directed deletion of MC1R in zygotes (Adopted from Xiao et al., 2019) Image caption: (A) Schematic diagram of sgRNA targeting the rabbit MC1R gene loci. The yellow rectangle represents the transmembrane domain of MC1R. Two sgRNA sequences, sgRNA1 (Sg1#) and sgRNA2 (Sg2#), are highlighted in green. Protospacer adjacent motif (PAM) sequences are presented in red with underline. Primers F and R are used for mutation detection in embryos. (B) Cytoplasmic injection of zygotes using the CRISPR/Cas9 system. Seven blastocysts are collected. Mutation detection in blastocyst by PCR. M, marker; numbers 1–7 represent different blastocysts used in this study. The number in red represents the positive embryos. Scale bar, 100 μm. (C) T-cloning and Sanger sequencing of the modified MC1R alleles in blastocysts. Wild-type sequence is shown at the top of the targeting sequence. Sequences of sgRNAs are marked in green, the PAM sequences are in red, insertions are highlighted in lowercase red letters, and deletions are designated by dashes. E: embryos; WT: ED allele; deletion: “−”; insertion: “+” (Adopted from Xiao et al., 2019) |
In another study, researchers sequenced the MC1R gene in several domestic rabbits and identified four alleles, including two wild-type alleles and two alleles with in-frame deletions. These deletions were associated with different coat colors, such as dominant black and recessive red. The study highlighted the role of MC1R gene mutations in determining coat color and provided a foundation for further genetic editing experiments (Fontanesi et al., 2006).
4.3 Epigenetic factors influencing coat color
Epigenetic modifications, such as DNA methylation, have been shown to influence coat color in domestic rabbits. A study on Rex rabbits compared genome-wide DNA methylation profiles in hair follicles between a Chinchilla group and a diluted Chinchilla group using whole-genome bisulfite sequencing (WGBS). The researchers identified 126 405 differentially methylated regions (DMRs) corresponding to 11 459 DMR-associated genes. These genes were primarily involved in developmental pigmentation and Wnt signaling pathways. The study provided evidence that inherited color dilution is associated with DNA methylation alterations in hair follicles, contributing to our understanding of the epigenetic regulation of rabbit pigmentation (Chen et al., 2020).
Another study focused on the melanophilin (MLPH) gene, which is associated with coat color dilution in rabbits. The researchers identified two variants in the MLPH gene: an acceptor splice site variant (c.111-5C>A) and a frameshift mutation (c.585delG). They analyzed the allelic segregation and functional effects of these variants on MLPH transcript stability. The study revealed that the c.585delG variant showed perfect association with the dilution phenotype and led to a significant decrease in MLPH transcript levels in rabbits with diluted coat color. These findings provided new insights into the molecular mechanisms underlying coat color dilution.
5 Case Study: Genetic Analysis of a Specific Rabbit Population
5.1 Background and selection of the case study population
The genetic basis of coat color variations in domestic rabbits (Oryctolagus cuniculus) has been a subject of extensive research due to its economic and breed characteristic importance. This case study focuses on the genetic analysis of coat color variations in a specific population of Chinese Rex rabbits. The Rex rabbit breed is known for its unique fur structure and a variety of coat colors, making it an ideal candidate for genetic studies on pigmentation.
The selection of the Chinese Rex rabbit population for this case study is based on several factors. The breed exhibits a wide range of coat colors, including castor, red, blue, chinchilla, otter, and black, which provides a diverse genetic pool for analysis (Yang et al., 2015). Previous studies have identified several candidate genes associated with coat color variations in rabbits, such as MC1R, MITF, TYR, TYRP1, MLPH, and ASIP, making it feasible to investigate these genes in the selected population (Chen et al., 2020; Xiao et al., 2019; Utzeri et al., 2001; Fontanesi et al., 2006; Jia et al., 2021; Zhang et al, 2023). Lastly, the economic significance of coat color in the Rex rabbit industry further justifies the selection of this population for genetic analysis (Zhang et al, 2023).
5.2 Results and Interpretation
The genetic analysis of the Chinese Rex rabbit population involved genome-wide association studies (GWAS) and sequencing of candidate genes known to influence coat color. Blood samples were collected from 250 Chinese Rex rabbits with six different coat colors, and genome sequencing was performed using a restriction site-associated DNA sequencing approach. A total of 91 546 single nucleotide polymorphisms (SNPs) were identified, distributed across 21 autosomes (Zhang et al, 2023).
The GWAS identified 24 significant SNPs located within a genomic region on chromosome 4 (OCU4), with the most significant SNP being OCU4:13,434,448 (p=1.31e-12). This region was found to be significantly associated with coat color in Chinese Rex rabbits, and the well-studied agouti signaling protein (ASIP) gene was located within this region (Figure 2) (Zhang et al, 2023). The ASIP gene is known to play a crucial role in determining coat color by regulating the distribution of eumelanin and pheomelanin pigments.
 Figure 2 Genome-wide association with coat colors of Chinese Rex rabbits (Adopted from Zhang et al., 2023). After testing all SNP effects with a mixed linear model (top panel), the most significant SNP (OCU4:13,434,448) was fitted as a covariate for re-testing of SNP effects (middle panel). Both significant SNPs (OCU4:13,434,448 and OCU4:11,344,946) were simultaneously fitted as covariates for re-testing of SNP effects (bottom panel). The dashed line represents the genome-wide significance threshold (Adopted from Zhang et al., 2023) |
In addition to the ASIP gene, variations in other pigmentation genes such as MC1R, MITF, TYR, TYRP1, and MLPH were also analyzed. For instance, the MC1R gene was found to have multiple alleles associated with different coat colors, including a 30-nucleotide in-frame deletion (c.304_333del30) associated with red/fawn/yellow coat colors and a 6-nucleotide in-frame deletion (c.280_285del6) associated with black coat colors (Fontanesi et al., 2006; Fontanesi et al., 2010; Xiao et al., 2019). Similarly, the TYR gene, which is responsible for albinism, exhibited several missense mutations associated with different coat color phenotypes, such as the albino, Himalayan, and chinchilla alleles (Utzeri et al., 2021).
Furthermore, the MLPH gene, which affects coat color dilution, was found to have two variants associated with the dilution phenotype: the c.111-5C>A variant and the c.585delG variant. The c.585delG variant showed a perfect association with the dilution phenotype, and a significant decrease in MLPH transcript levels was observed in rabbits with diluted coat colors.
5.3 Implications for Breeding Programs
The findings from this genetic analysis have significant implications for breeding programs aimed at improving coat color traits in domestic rabbits. The identification of specific genetic variants associated with coat color provides valuable molecular markers that can be used in selective breeding programs to achieve desired coat colors.
For instance, the identification of the ASIP gene as a major determinant of coat color variation in Chinese Rex rabbits allows breeders to select for specific alleles to produce rabbits with preferred coat colors. Similarly, the variations in the MC1R and TYR genes can be used to predict and select for specific coat color phenotypes, such as red/fawn/yellow or albino rabbits (Fontanesi et al., 2006; Fontanesi et al., 2010; Yang et al., 2015; Xiao et al., 2019; Utzeri et al., 2021). Moreover, the association of the MLPH gene with coat color dilution provides a genetic basis for selecting rabbits with diluted coat colors, which are often preferred in the rabbit fur industry. By selecting for the c.585delG variant, breeders can produce rabbits with consistent and desirable coat color dilution.
In addition to improving coat color traits, the genetic information obtained from this study can also be used to maintain genetic diversity within rabbit populations. By understanding the genetic basis of coat color variations, breeders can avoid inbreeding and ensure the long-term sustainability of rabbit breeds. The genetic analysis of coat color variations in the Chinese Rex rabbit population provides a comprehensive understanding of the genetic mechanisms underlying coat color traits. This knowledge can be effectively applied in breeding programs to enhance the economic and aesthetic value of domestic rabbits.
6 Applications of Coat Color Genetics in Rabbit Breeding
6.1 Selection for desirable traits in breeding
The genetic analysis of coat color variations in domestic rabbits has significant implications for the selection of desirable traits in breeding programs. Coat color is not only an aesthetic trait but also an economic one, influencing the market value of rabbits. The identification of specific genes and their polymorphisms associated with coat color can provide breeders with molecular markers to select for preferred traits more efficiently.
For instance, the study on the polymorphism of MC1R, MITF, TYR, TYRP1, and MLPH genes in four Chinese native rabbit breeds revealed 14 genetic variants that play a crucial role in determining coat color (Jia et al., 2021). These genetic markers can be used to predict and select for specific coat colors in breeding programs, ensuring the propagation of desirable traits. Similarly, the use of single-nucleotide polymorphism (SNP) markers from genotyping-by-sequencing data has identified genes such as ASIP, MITF, and KIT that are associated with fur color (Li et al., 2022). These markers can be integrated into breeding strategies to enhance the selection process, making it more precise and efficient.
Moreover, the identification of these genetic markers allows for the development of breeding programs that can maintain or enhance the genetic diversity of rabbit populations. By understanding the genetic basis of coat color, breeders can avoid inbreeding and ensure the health and vitality of rabbit populations. This approach not only improves the aesthetic and economic value of rabbits but also contributes to the overall sustainability of rabbit breeding practices.
6.2 Ethical considerations in coat color manipulation
While the manipulation of coat color through genetic selection offers numerous benefits, it also raises several ethical considerations. The primary concern is the welfare of the animals involved in breeding programs. Selective breeding for specific traits, such as coat color, can sometimes lead to unintended consequences, including health issues and reduced genetic diversity.
Ethical breeding practices must prioritize the well-being of the animals. This includes ensuring that the selection for coat color does not compromise other important traits, such as health, behavior, and overall fitness. For example, the study on the genetic variations in pigmentation genes highlights the importance of considering the broader genetic context when selecting for coat color (Jia et al., 2021). Breeders must be cautious not to inadvertently select for deleterious alleles that could negatively impact the health of the rabbits.
Additionally, there is a need for transparency and informed consent in breeding practices. Breeders should provide accurate information about the genetic modifications and their potential implications to stakeholders, including consumers and regulatory bodies. This transparency helps build trust and ensures that ethical standards are upheld in the breeding industry. The use of genetic technologies in breeding should be guided by ethical frameworks that consider the long-term impacts on rabbit populations and ecosystems. This includes evaluating the potential risks and benefits of genetic manipulation and ensuring that breeding practices align with principles of animal welfare and environmental sustainability.
6.3 Future prospects in rabbit breeding
The future of rabbit breeding is poised to benefit significantly from advancements in genetic research and technology. The identification of genetic markers associated with coat color and other traits opens up new possibilities for precision breeding. This approach allows breeders to select for specific traits with greater accuracy, reducing the time and resources required for traditional breeding methods.
One promising area of research is the use of genome-wide association studies (GWAS) to identify additional genetic markers linked to desirable traits. The study on SNP markers from genotyping-by-sequencing data provides a foundation for such research, highlighting the potential of GWAS to uncover new genetic variants associated with coat color and other economically important traits (Li et al., 2022). These findings can be integrated into breeding programs to enhance the selection process and improve the overall quality of rabbit breeds.
Another exciting prospect is the application of gene editing technologies, such as CRISPR-Cas9, in rabbit breeding. These technologies offer the potential to directly modify specific genes associated with coat color, allowing for precise control over the genetic makeup of rabbits. However, the use of gene editing must be approached with caution, considering the ethical and regulatory implications.
In addition to genetic technologies, advancements in reproductive technologies, such as in vitro fertilization (IVF) and embryo transfer, can further enhance rabbit breeding programs. These technologies enable the rapid propagation of desirable traits and the preservation of genetic diversity. Combined with genetic markers, reproductive technologies can accelerate the development of superior rabbit breeds with optimal coat color and other traits.
7 Challenges and Future Directions
7.1. Limitations of current genetic studies
Current genetic studies on coat color variations in domestic rabbits have provided significant insights but also face several limitations. One major limitation is the focus on a limited number of genes. For instance, studies have primarily concentrated on genes like MC1R, MITF, TYR, TYRP1, and MLPH, which are known to influence pigmentation (Fontanesi et al., 2006; Jia et al., 2021; Utzeri et al., 2021). While these genes are crucial, the genetic architecture of coat color is likely more complex, involving additional genes and regulatory elements that have not yet been fully explored.
Another limitation is the sample size and diversity. Many studies have been conducted on specific breeds or populations, which may not capture the full genetic diversity of domestic rabbits. For example, the study on Chinese Rex rabbits involved 250 individuals but was limited to six coat color varieties (Li et al., 2022). Similarly, research on the TYR gene included only 25 rabbits from 11 domestic breeds and a wild population from Sardinia (Utzeri et al., 2021). These sample sizes, while informative, may not be sufficient to generalize findings across all rabbit breeds.
Moreover, the methodologies used in these studies, such as genome-wide association studies (GWAS) and DNA sequencing, have their own set of limitations. GWAS, for instance, can identify associations but not causations, and the resolution of these studies is often limited by the density of the SNP markers used (Alves et al., 2015; Li et al., 2022) Additionally, while DNA sequencing can identify genetic variants, it does not provide information on the functional impact of these variants, which requires further experimental validation.
7.2 Emerging technologies and their potential impact
Emerging technologies hold great promise for overcoming some of the limitations of current genetic studies. One such technology is whole-genome sequencing (WGS), which provides a comprehensive view of the genetic makeup of an organism. WGS can identify both common and rare variants, offering a more complete picture of the genetic factors influencing coat color (Chen et al., 2020). Additionally, advances in bioinformatics tools and computational power allow for more sophisticated analyses of these large datasets, potentially uncovering new genetic associations and interactions.
Another promising technology is CRISPR-Cas9, a powerful tool for genome editing. CRISPR-Cas9 can be used to create targeted mutations in specific genes, allowing researchers to study their functional impact directly. This technology could be particularly useful for validating the roles of candidate genes identified in GWAS and sequencing studies (Fontanesi et al., 2013). For example, if a specific SNP in the MC1R gene is associated with a particular coat color, CRISPR-Cas9 could be used to introduce this SNP into a different genetic background to observe its effect.
Epigenetic studies are also emerging as a crucial area of research. DNA methylation and histone modifications can influence gene expression without altering the underlying DNA sequence. Recent studies have begun to explore the role of epigenetics in coat color variations, revealing that DNA methylation patterns can differ significantly between rabbits with different coat colors (Chen et al., 2020). These findings suggest that epigenetic modifications could be an important regulatory mechanism in pigmentation, and further research in this area could provide new insights into the genetic control of coat color.
7.3 Areas for further research
Several areas warrant further research to advance our understanding of coat color variations in domestic rabbits. One important area is the identification and functional characterization of additional genes involved in pigmentation. While genes like MC1R, MITF, and TYR have been well-studied, other genes and regulatory elements likely play significant roles in determining coat color. High-throughput sequencing technologies and functional genomics approaches, such as RNA sequencing and chromatin immunoprecipitation sequencing (ChIP-seq), could be employed to identify these additional genetic factors (Fontanesi et al., 2006; Jia et al., 2021; Zhang et al., 2023).
Another area for further research is the study of gene-environment interactions. Coat color can be influenced by environmental factors such as temperature and light exposure, and understanding how these factors interact with genetic determinants could provide a more comprehensive understanding of pigmentation. For example, a study on wild rabbits in different climatic regions of Australia found significant changes in coat color over time, suggesting that environmental selection pressures can shape coat color variations.
Additionally, more research is needed on the epigenetic regulation of coat color. Studies have shown that DNA methylation patterns can differ between rabbits with different coat colors, but the specific mechanisms by which these epigenetic modifications influence pigmentation are not yet fully understood (Zhang et al., 2023). Further research in this area could involve genome-wide methylation studies and the use of epigenetic editing tools to manipulate methylation patterns and observe their effects on coat color.
Finally, there is a need for more comprehensive and diverse studies that include a wider range of rabbit breeds and populations. This would help to capture the full genetic diversity of domestic rabbits and provide more generalizable findings. Collaborative efforts and the establishment of large, multi-breed cohorts could facilitate such studies and lead to a deeper understanding of the genetic basis of coat color variations in domestic rabbits (Alves et al. , 2015; Utzeri et al., 2021; Li et al., 2022).
8 Concluding Remarks
The genetic analysis of coat color variations in domestic rabbits has revealed significant insights into the underlying genetic mechanisms. Key findings include the identification of multiple genetic variants in pigmentation genes such as MC1R, MITF, TYR, TYRP1, and MLPH, which play crucial roles in determining rabbit coat color1. Genome-wide association studies (GWAS) have pinpointed significant single nucleotide polymorphisms (SNPs) associated with coat color, particularly highlighting the agouti signaling protein (ASIP) gene as a major determinant in Chinese Rex rabbits. Additionally, specific mutations in the MC1R gene have been linked to various coat color phenotypes, including dominant black, recessive red, and Japanese brindling. The TYR gene has also been implicated in albinism and other color variations through several missense mutations. Furthermore, the use of CRISPR/Cas9 technology has demonstrated the potential to create novel coat colors by inducing targeted mutations in the MC1R gene. Epigenetic factors, such as DNA methylation, have been shown to influence coat color dilution, adding another layer of complexity to the genetic regulation of pigmentation.
The findings from these studies have significant implications for future research and breeding strategies. The identification of specific genetic variants and their associations with coat color phenotypes provides valuable molecular markers that can be used in selective breeding programs to achieve desired coat colors in domestic rabbits. The potential of gene editing technologies, such as CRISPR/Cas9, to create new coat color phenotypes opens up exciting possibilities for the rabbit breeding industry, allowing for the development of novel and economically valuable traits. Additionally, understanding the role of epigenetic modifications in coat color variation suggests that future research should explore the interplay between genetic and epigenetic factors to fully elucidate the mechanisms of pigmentation6. This knowledge can be leveraged to enhance breeding strategies, ensuring the production of rabbits with specific and consistent coat colors.
The role of genetics in coat color variation in domestic rabbits is both intricate and multifaceted. The studies reviewed highlight the significant contributions of various genes and their polymorphisms to the diverse coat color phenotypes observed in rabbit populations. The integration of genetic, epigenetic, and gene editing approaches provides a comprehensive understanding of the mechanisms driving coat color variation. This integrated approach not only advances our scientific knowledge but also offers practical applications in rabbit breeding, enabling the development of rabbits with specific and desirable coat colors. As research continues to uncover the complexities of genetic regulation, the potential for innovative breeding strategies and the creation of novel phenotypes will undoubtedly expand, benefiting both the scientific community and the rabbit breeding industry.
By leveraging the insights gained from these genetic studies, breeders can make informed decisions to enhance the aesthetic and economic value of domestic rabbits, ultimately contributing to the advancement of the field of animal genetics and breeding.
Acknowledgements
Author would like to express our gratitude to the two anonymous peer reviewers for their critical assessment and constructive suggestions on our manuscript.
Conflict of Interest Disclosure
Author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Alves J.M., Carneiro M., Afonso S., Lopes S., Garreau H., Boucher S., Allain D., Queney G., Esteves P.J., Bolet G., and Ferrand N., 2015, Levels and patterns of genetic diversity and population structure in domestic rabbits, PLoS One, 10(12): e0144687.
https://doi.org/10.1371/journal.pone.0144687
Ballan M., Bovo S., Bertolini F., Schiavo G., Schiavitto M., Negrini R., and Fontanesi L., 2023, Population genomic structures and signatures of selection define the genetic uniqueness of several fancy and meat rabbit breeds, Journal of Animal Breeding and Genetic, 140(6):663-678.
https://doi.org/10.1111/jbg.12818
PMID: 37435689
Chen Y., Hu S.S., Liu M., Zhao B.H., Yang N., Li J., Chen Q., Zhou J., Bao G., and Wu X., 2020, Analysis of genome DNA methylation at inherited coat color dilutions of rex rabbits, Frontiers in Genetics, 11: 603528.
https://doi.org/10.3389/fgene.2020.603528
PMID: 33552123 PMCID: PMC7859435
Chhotaray S., Panigrahi M., Bhushan B., Gaur G., Dutt T., Mishra B., and Singh R., 2021, Genome-wide association study reveals genes crucial for coat color production in Vrindavani cattle, Livestock Science, 247: 104476.
https://doi.org/10.1016/J.LIVSCI.2021.104476
Carneiro M., Afonso S., Geraldes A., Garreau H., Bolet G., Boucher S., Tircazes A., Queney G., Nachman M.W., and Ferrand N., 2011, The genetic structure of domestic rabbits, Molecular Biology and Evolution, 28(6): 1801-1816.
https://doi.org/10.1093/molbev/msr003
PMID: 21216839 PMCID: PMC3695642
Demars J., Iannuccelli N., Utzeri V.J., Auvinet G., Riquet J., Fontanesi L., and Allain D., 2018, New insights into the melanophilin (MLPH) gene affecting coat color dilution in rabbits, Genes, 9(9): 430.
https://doi.org/10.3390/genes9090430
PMID: 30142960 PMCID: PMC6162760
Dorożyńska K., and Maj D., 2020, Rabbits-their domestication and molecular genetics of hair coat development and quality, Animal Genetics, 52(1): 10-20.
https://doi.org/10.1111/age.13024
PMID: 33216407
Fontanesi L., 2021, Rabbit genetic resources can provide several animal models to explain at the genetic level the diversity of morphological and physiological relevant traits, Applied Sciences, 11(1): 373.
https://doi.org/10.3390/app11010373
Fontanesi L., Scotti E., Allain D., and Dall'Olio S., 2014, A frameshift mutation in the melanophilin gene causes the dilute coat colour in rabbit (Oryctolagus cuniculus) breeds, Animal genetics, 45(2): 248-255.
https://doi.org/10.1111/age.12104
PMID: 24320228
Fontanesi L., Scotti E., Colombo M., Allain D., Deretz S., Dall'Olio S., Russo V., and Oulmouden A., 2013, Investigation of the premelanosome protein (PMEL or SILV ) gene and identification of polymorphism excluding it as the determinant of the dilute locus in domestic rabbits (Oryctolagus cuniculus), Archives Animal Breeding, 56(1): 42-49.
https://doi.org/10.7482/0003-9438-56-005
Fontanesi L., Scotti E., Colombo M., Beretti F., Forestier L., Dall'Olio S., Deretz S., Russo V., Allain D., and Oulmouden A., 2010, A composite six bp in-frame deletion in the melanocortin 1 receptor (MC1R) gene is associated with the Japanese brindling coat colour in rabbits (Oryctolagus cuniculus), BMC Genetics, 11: 59.
https://doi.org/10.1186/1471-2156-11-59
PMID: 20594318 PMCID: PMC3236303
Fontanesi L., Tazzoli M., Beretti F., and Russo V., 2006, Mutations in the melanocortin 1 receptor (MC1R) gene are associated with coat colours in the domestic rabbit (Oryctolagus cuniculus), Animal Genetics, 37(5): 489-493.
https://doi.org/10.1111/J.1365-2052.2006.01494.X
PMID: 16978179
Jia X.B., Ding P., Chen S., Zhao S., Wang J., and Lai S., 2021, Analysis of MC1R, MITF, TYR, TYRP1, and MLPH genes polymorphism in four rabbit breeds with different coat colors, Animals, 11(1): 81.
https://doi.org/10.3390/ani11010081
PMID: 33466315 PMCID: PMC7824738
Hartl G., and Höger H., 1986, Biochemical variation in purebred and crossbred strains of domestic rabbits (Oryctolagus cuniculus L.), Genetical Research, 48(1): 27-34.
https://doi.org/10.1017/S0016672300024629
PMID: 33466315 PMCID: PMC7824738
Li C.Y., Li Y., Zheng J., Guo Z., Mei X., Lei M., Ren Y., Zhang X., Zhang C.X., Yang C., Tang L., Ji Y., Yang R., Yu J.F., Xie X., and Kuang L.D., 2022, Trait analysis in domestic rabbits (Oryctolagus cuniculus f. domesticus) using SNP markers from genotyping-by-sequencing data, Animals, 12(16): 2052.
https://doi.org/10.3390/ani12162052
PMID: 36009642 PMCID: PMC9404428
Ludwig A., Pruvost M., Reissmann M., Benecke N., Brockmann G., Castaños P., Cieslak M., Lippold S., Llorente L., Malaspinas A., Slatkin M., and Hofreiter M., 2009, Coat color variation at the beginning of horse domestication, Science, 324: 485-485.
https://doi.org/10.1126/science.1172750
Qanbari S., Pausch H., Jansen S., Somel M., Strom T.M., Fries R., Nielsen R., and Simianer H., 2014, Classic selective sweeps revealed by massive sequencing in cattle, PLoS Genetics, 10(2): e1004148.
https://doi.org/10.1371/journal.pgen.1004148
PMID: 24586189 PMCID: PMC3937232
Qin L., Wang W., Shi L., Wan X., Yan X., Weng Q., and Wu X., 2016, Transcriptome expression profiling of fur color formation in domestic rabbits using Solexa sequencing, Genetics and molecular research GMR, 15(2): 15027413.
https://doi.org/10.4238/gmr.15027413
Stodart E., 1965, A study of the biology of the wild rabbit in climatically different regions in eastern Australia, III, Some data on the evolution of coat colour, Wildlife Research, 10: 73-82.
Utzeri V., Ribani A., and Fontanesi L., 2014, A premature stop codon in the TYRP1 gene is associated with brown coat colour in the European rabbit (Oryctolagus cuniculus), Animal Genetics, 45(4): 600-603.
https://doi.org/10.1111/age.12171
Utzeri V., Ribani A., Schiavo G., and Fontanesi L., 2021, Describing variability in the tyrosinase (TYR) gene, the albino coat colour locus, in domestic and wild European rabbits, Italian Journal of Animal Science, 20: 181-187.
https://doi.org/10.1080/1828051X.2021.1877574
Vašíčková K., Ondruška Ľ., Baláži A., Parkányi V., and Vašíček D., 2016, Genetic characterization of Nitra rabbits and Zobor rabbits, Slovak Journal of Animal Science, 49(3): 104-111.
Fang M.Y., Larson G., Ribeiro H.S., Li N., and Andersson L., 2009, Contrasting mode of evolution at a coat color locus in wild and domestic pigs, PLoS Genetics, 5(1): e1000341.
https://doi.org/10.1371/journal.pgen.1000341
PMID: 19148282 PMCID: PMC2613536
Xiao N., Li H.L., Shafique L., Zhao S., Su X., Zhang Y., Cui K., Liu Q.Y., and Shi D.S., 2019, A novel pale-yellow coat color of rabbits generated via MC1R mutation with CRISPR/Cas9 system, Frontiers in Genetics, 10: 875.
https://doi.org/10.3389/fgene.2019.00875
PMID: 31620174 PMCID: PMC6759607
Yang C.J., Ge J., Chen S.J., Liu Y.J., Chen B.J., and Gu Z.L., 2015, Sequence and gene expression analysis of the agouti signalling protein gene in rex rabbits with different coat colours, Italian Journal of Animal Science, 14(3): 3810.
https://doi.org/10.4081/ijas.2015.3810
Zhang K., Wang G.Z., Wang L.H., Wen B., Fu X.C., Liu N., Yu Z.J., Jian W.S., Guo X., Liu H., and Chen S.Y., 2023, A genome-wide association study of coat color in Chinese rex rabbits, Frontiers in Veterinary Science, 10: 1184764.
https://doi.org/10.3389/fvets.2023.1184764
PMID: 37655262 PMCID: PMC10467280
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xinghao Li
. Jia Xuan
Related articles
. Coat color
. Domestic rabbits
. MC1R gene
. SNP markers
. Pigmentation genetics
Tools
. Post a comment


